Unsupervised Learning for Plant Disease Detection
Data
Our final dataset includes 5 sources:
- Plant Village
- Tomato Dataset
- PlantDoc Dataset
- Bing Image Search
- GAN Images

Data Augmentation:
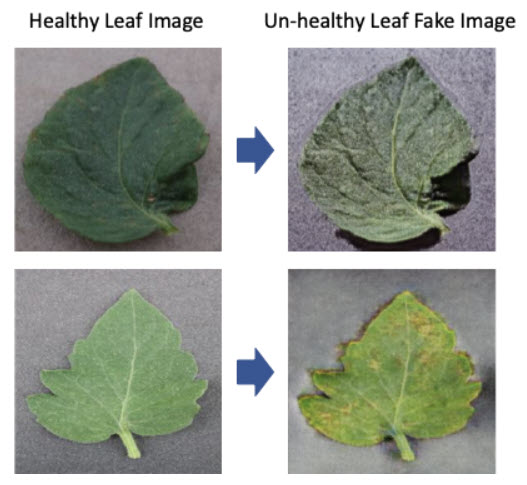
The image augmentation algorithm was employed to increase the number of certain images due to the unbalanced dataset and to increase the robustness of the model. The conventional augmentation schemes like average blur, rotation, affine, translate, cutout, resize, etc together with a generative adversarial network were employed. Generative adversarial network (GAN) is a machine learning model that can learn to mimic a given distribution of data. GAN consists of two separate neural network models: a generative model and a discriminative model. The discriminative model learns how to classify input to its class and in GAN, it is a binary classifier to determine whether a given image is a real image from the real dataset or an artificially created image. The generative model learns the distribution of training data and in GAN, it is a data generator to transform input values into images through a deconvolutional neural network. The goal of the generator is to generate the new images which the discriminator cannot distinguish from real images and the goal of the discriminator is to correctly tell real images apart from fake images created by the generator. Since the first publication of GAN [Ian Goodfellow NIPS, 2014], many types of GAN have been developed. In this project, the CycleGAN, one of conditional GAN models, was employed to augment the image data for disease detection and it is the image-to-image translation algorithm without need of paired examples of transformation from source to target domain [https://junyanz.github.io/CycleGAN/]. It can transform the image from one domain to another domain without a one-to-one mapping between the source and target domain. For example, it can generate an unhealthy leaf image from the healthy leaf image domain and the healthy leaf image is also generated from the unhealthy leaf image domain. Our data set was very unbalanced. It means the number of images of the specific classes is much smaller than others and it can cause the bias in the model. Using CycleGAN, the images of the specific classes were generated and fed to the model to resolve the unbalanced issue.
Model Training:
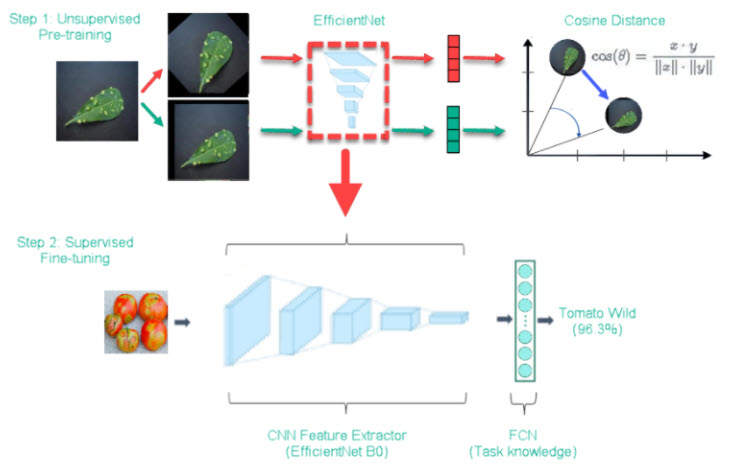
We follow the unsupervised training framework in SimCLR (A Simple Framework for Contrastive Learning of Visual Representations) with a few modifications. In step 1, we used EfficientNet as base mode and trained on 35,000 images that are not part of plant disease dataset. In step 2, we finetune the base model with labeled data.
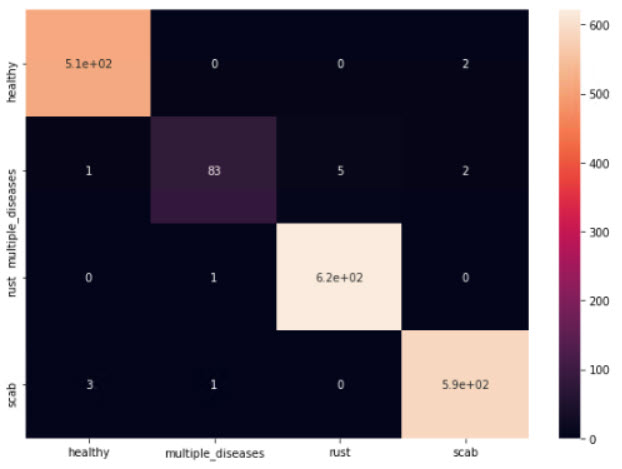
This framework appears to be working for our toy dataset with 4 classes (Plant Pathology 2020 FGVC7):
Error Analysis:
We applied Grad-CAM on test images to visualize where our model focuses on in the image during inference. Below is one of the examples of original image (left) and Grad-Cam image overlay (right):

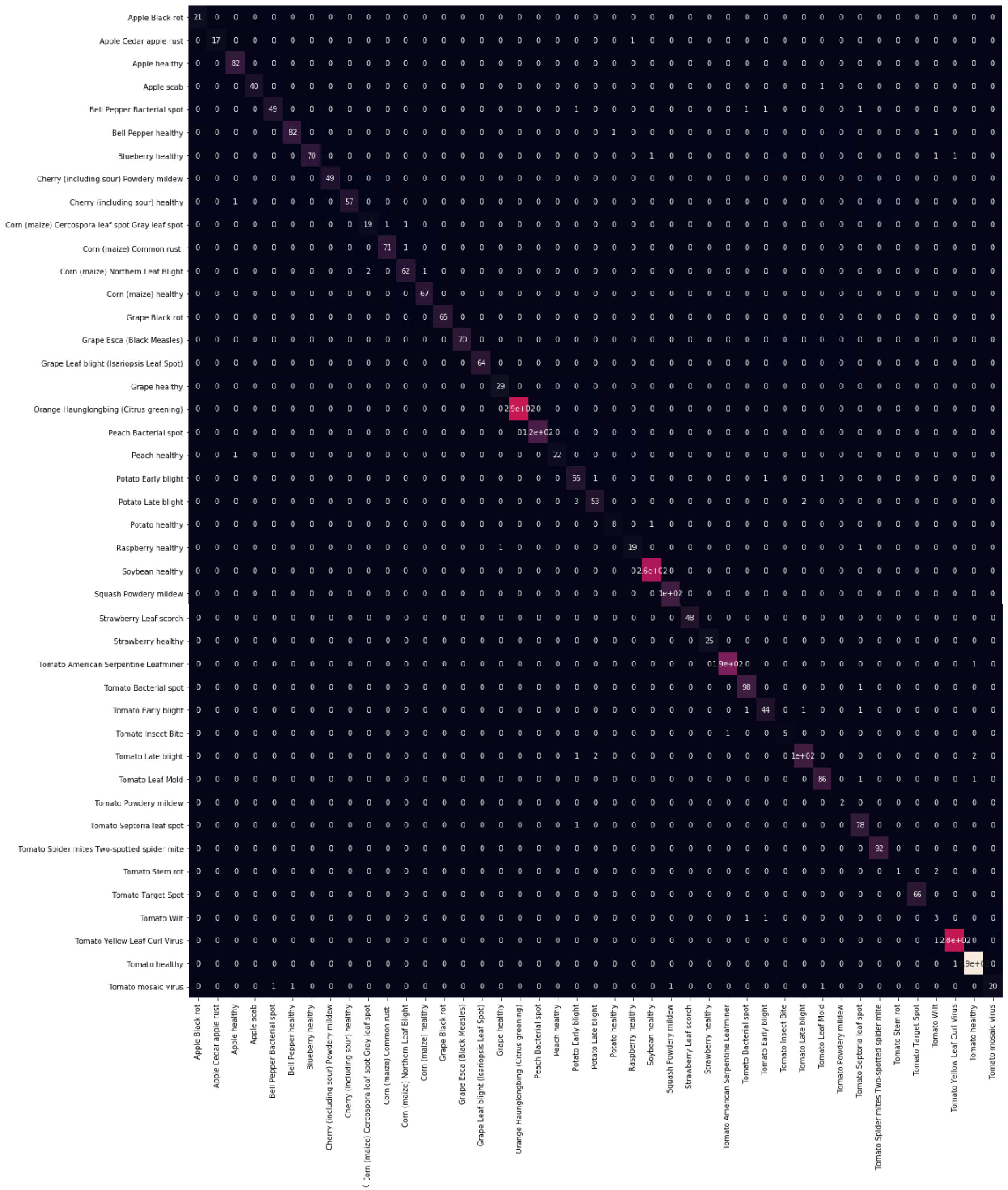
Final Confusion Matrix
Knowledge Base:
To provide users with follow up information, a knowledge base of plant disease is built. It’s used in both web prediction and phone app to provide more detail explanation of the plant disease predicted. Or it could be used as an QnA chatbox through channels like Slack, Teams or emails. Knowledge base is base Microsoft Open Source Bot Framework connecting to backend knowledge base.
Phone App:
https://github.com/louis-li/PhoneApp